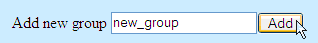Help
|
|
Main page
Algorithm & format requirements
Help
Contact us
|
|
|
Using stand-alone SDPfox
From our site you can download stand-alone SDPfox,
and example alignment.
To get help, run "java -jar SDPfox.jar". Java version 1.6.0_17 or later is required.
Web interface
Alignment
To add a new specificity group, type in the group name and press "Add"
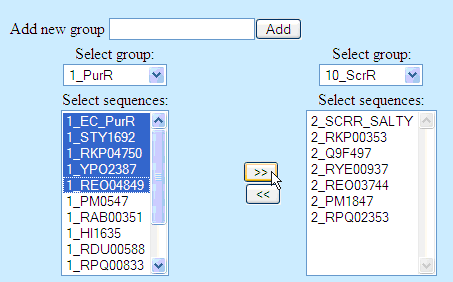
To change composition of specificity groups, select two groups: first, from which to remove
sequences, and second, to which to add sequences. Select sequences and press one of the arrow
buttons:
Click name of a group or a sequence to get detailed information on a position (SDP)
SDPgroup
For a constructed grouping, it is possible to run SDPgroup, re-randomize grouping or move
sequences according to the best weights (see algorithm) under the link 'SDPgroup'.
There, the user is requested to select a method and press "GO".
To randomize grouping, set number of specificity groups and number of sequences into each group:

After the randomization is done, PSSM group weights are calculated for each sequence and those
that have a higher weight for a group other than the initial group are marked red. These sequences
are likely to be re-grouped after SDPgroup is performed.
|
|