

The program calculates the correlation between each pair of columns [i, j]: one from the proteins alignment, and the other from the site alignment.
As a measure of correlation, the mutual information is used:
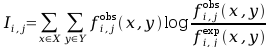 where:
where:
| X, Y | Arrays of 20 aminoacids and 4 nucleotides respectively. |
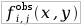 | Observed frequency of aminoacid x being in position i and nucleotide y being in position j. |
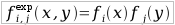 | Expected frequency with the hypothesis of absence of correlations between columns. Calculated as frequency of aminoacid x in column i multiplied by frequency of nucleotide y in column j |
Large difference of the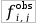 and the
expected distribution
and the
expected distribution 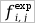 is reflected in high mutual information value
is reflected in high mutual information value
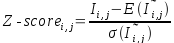 where:
where: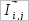 - is the distribution of the mutual information for random (non-correlated)
pairs of columns with the same aminoacid and nucleotide compositions as in [i, j] pair.
- is the distribution of the mutual information for random (non-correlated)
pairs of columns with the same aminoacid and nucleotide compositions as in [i, j] pair.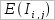 and
and 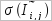 are the mean and the standard deviation, respectively.
are the mean and the standard deviation, respectively.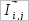 , 10,000 random pairs of columns are generated and mutual informtion values are calculeted for them.
, 10,000 random pairs of columns are generated and mutual informtion values are calculeted for them.