Server for multiple alignment of RNA sequences
General Algorithm
- Given word length and max number of mismatches
- Use MEME algorithm for define conserved blocks
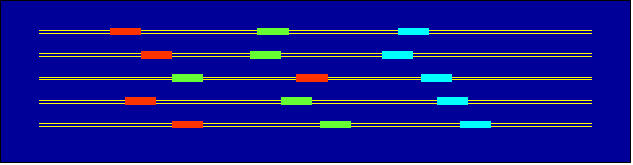
- Apply the Dynamic programming algorithm to create chain of the blocks
- Refine the chain using combined profile for chain of blocks.
The variation of distances between blocks produces gap penalty.
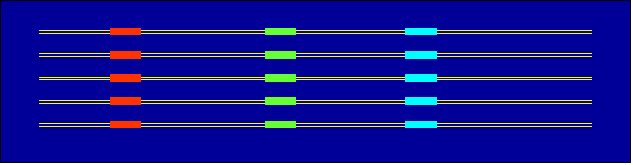
- Reduce word length and number of mismatches and repeat the procedure
for spacer between the blocks. Repeat the procedure until word size >3

-
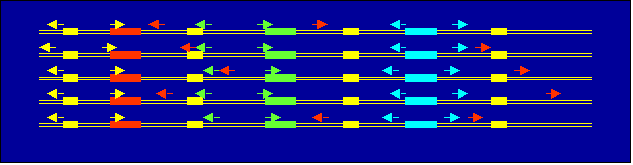
Find potential helices on the sequences with given perameters (loop soze and energy)
-
Create helix profile and apply MEME-like iterative procedure to find
sets of helices that consistent lokated reative to conserved blocks
Author:
Andrey Mironov
Department of Department of Bioengineering and Bioinformatics, MSU, Moscow, Russia
Mail to Andrey Mironov



